Our Programs
Targeted Protein Inhibition
Targeting cancer by inhibiting the activity of LSD1, a protein that can regulate gene expression
IN BRIEF
Targeted protein inhibition (TPI) of lysine-specific demethylase 1 (LSD1) activity is of great interest to cancer researchers. LSD1 is a protein involved in maintaining balanced gene expression. Gene expression is the process in which information stored in a gene is used to create proteins. Imbalanced gene expression can create a cancerous tumor. LSD1 has two key functions: removing specific chemical groups from proteins and bringing proteins together to form complexes that affect gene expression.
LSD1 is a promising inhibition target because
- It can both promote and block gene expression
- When it acts incorrectly, it can lead to cancer development and progression
- It has broad association in some cancers with various other gene expression–regulating proteins
- Inhibiting its functions may have anti-tumor activity across various cancer types, including solid tumors and hematologic cancers
IN DEPTH
Understanding epigenetically driven cancers begins with the genome, the complete set of genetic instructions in a cell. Genetic code is like a recipe, with proteins as the finished product. The genome is the cookbook.
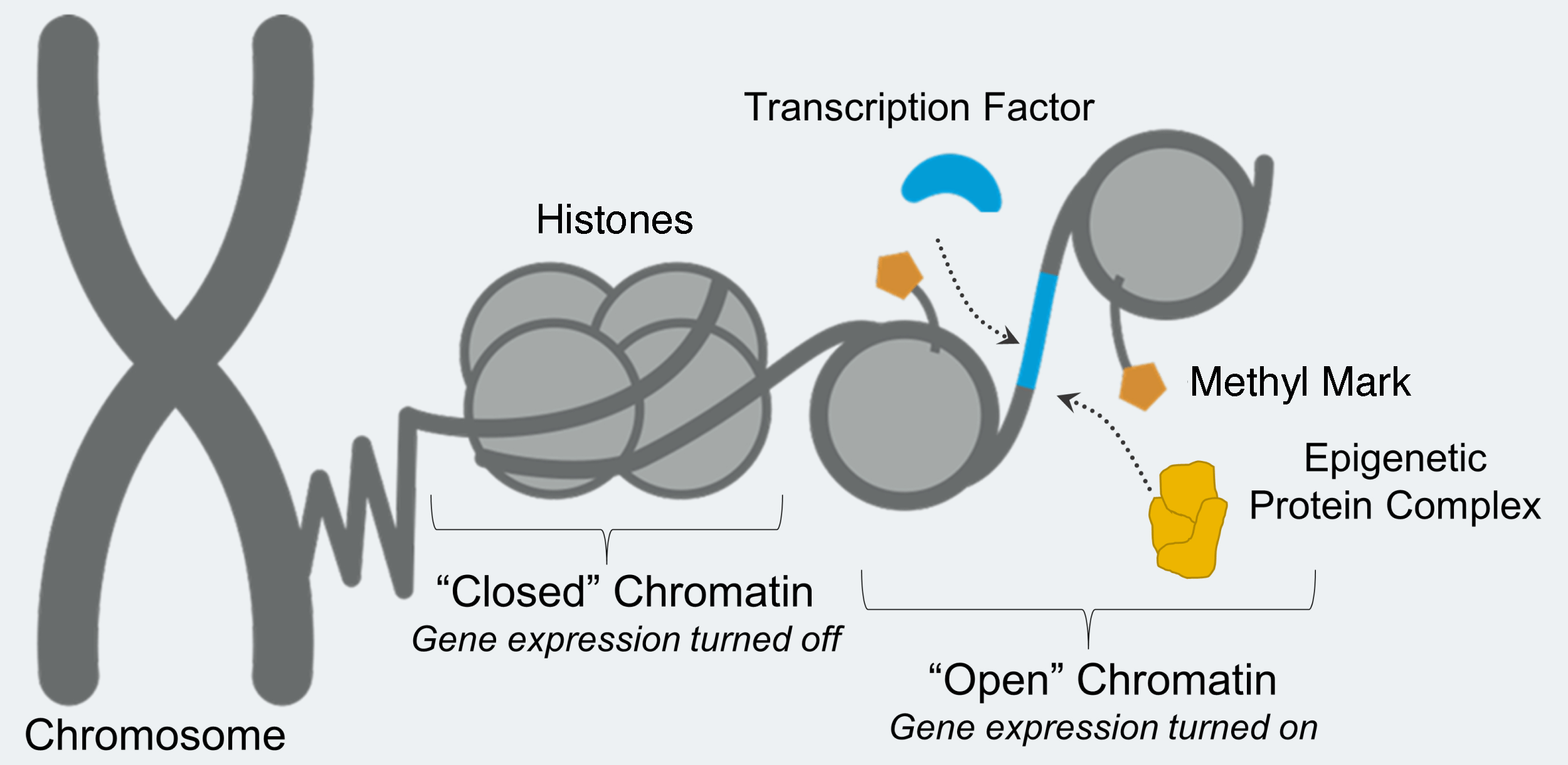
Sometimes changes can occur in DNA interactions without changing the DNA itself, like substituting an ingredient when preparing a recipe. Epigenetics is the complete set of changes that occur on DNA-associated proteins and ultimately affect the time, location, and duration of gene expression. When any of these changes occurs in the wrong place in the body or at the wrong time, it can lead to development of numerous diseases, including cancer.
LSD1 removes chemical groups from proteins associated with DNA, called histones. The location and number of chemical groups on histones determines whether the DNA can be accessed by proteins that will read that DNA and begin the process of making proteins. Accessible DNA is referred to as “open,” and inaccessible DNA is referred to as “closed.”
LSD1 Lysine-Specific Demethylase 1
LSD1: an epigenetic enzyme that can act incorrectly and lead to cancer development and progression
Because of this connection between LSD1 and cancer, researchers are greatly interested in developing drugs that can inhibit LSD1’s activity to treat both solid tumors and hematologic cancers.
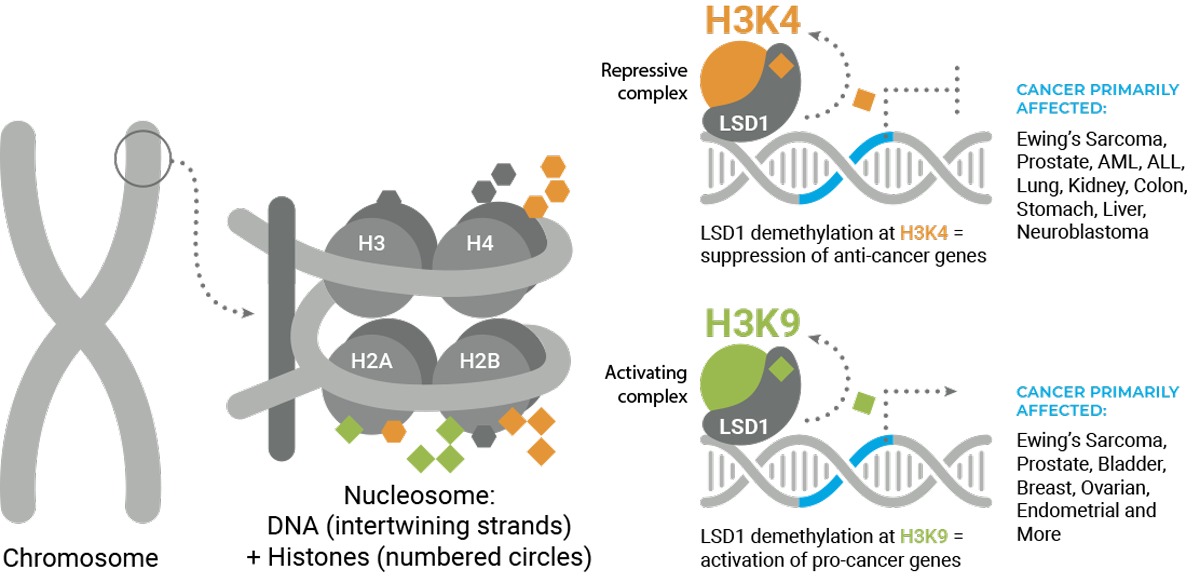
LSD1’s demethylation activity removes chemical groups, methyl marks, from protein-DNA structures called nucleosomes. Nucleosomes consist of DNA wrapped around structural proteins called histones. Histones, like most proteins, contain the amino acid lysine, which is particularly important for chemical modification.
The presence or absence of methyl marks determines whether DNA is open, available for transcription of genes, or closed, blocking gene expression. LSD1 specifically erases methyl marks at H3K4 and H3K9.
LSD1 closes DNA by removing the methyl mark at H3K4 and opens DNA by removing the methyl mark at H3K9. LSD1 can therefore both promote and block gene expression, which makes it unique as an epigenetic modifier. Finally, each lysine can have up to three methyl marks, and LSD1 is capable of removing only the first two marks. The number of methyl marks can also have an effect on gene expression.
Example of how LSD1’s scaffolding properties can drive cancer growth in Ewing sarcoma
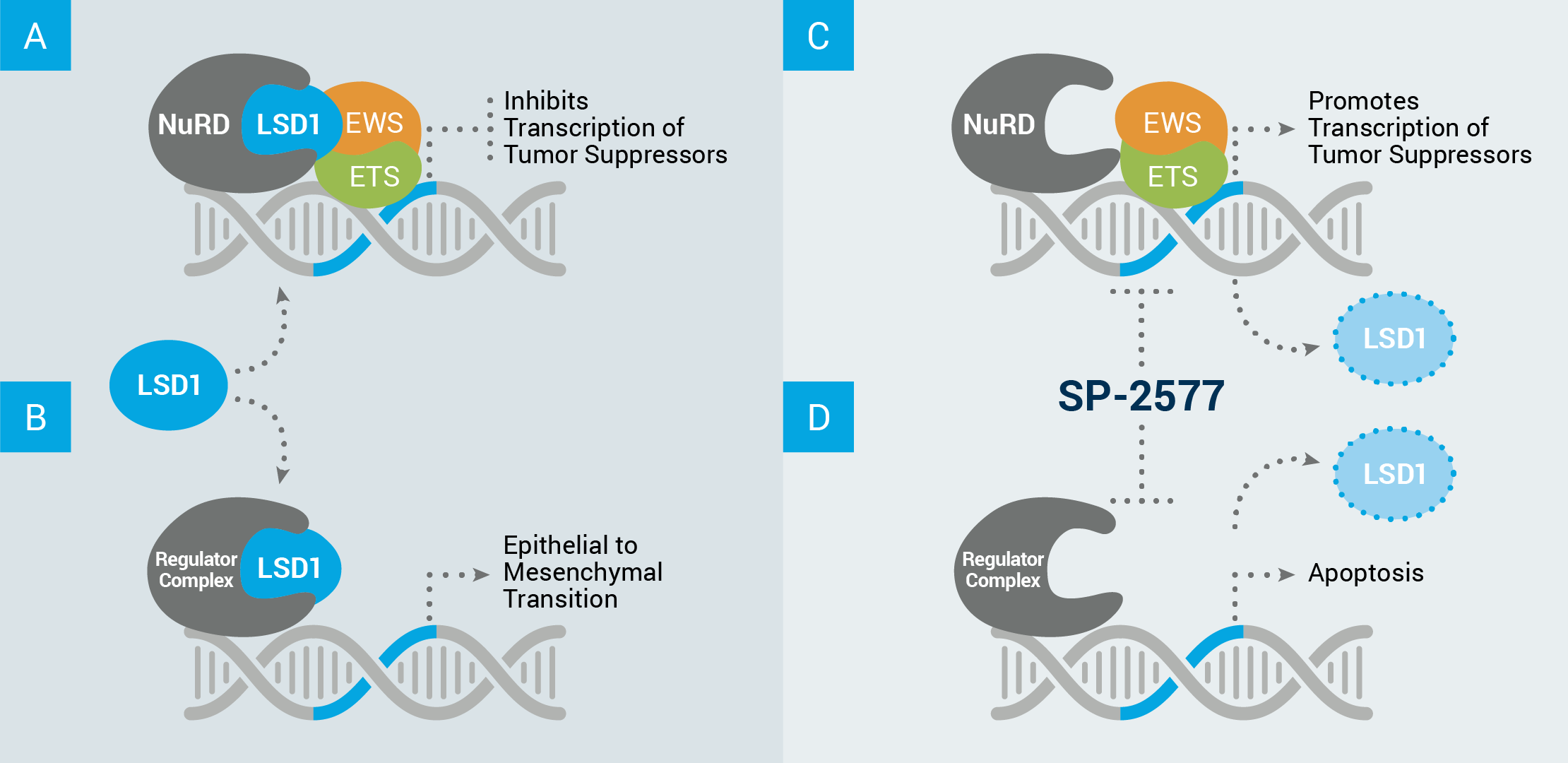
(A) LSD1 acts in complex with other proteins, such as NuRD, to inhibit the transcription of tumor suppressor genes.
(B) LSD1 can also interact with other proteins to disrupt the equilibrium of healthy cells and drive expression of a stem cell–like phenotype in which cells designed to perform a tissue-specific function (differentiated cells) revert to an undefined state of constant growth and invasion of nearby tissues.
Our lead compound, seclidemstat (SP-2577), inhibits LSD1’s scaffolding properties, which (C) allows for the transcription of tumor suppressor genes, and/or (D) prevents transcription relating to self-renewal, and can cause cell apoptosis (cell death).
ABBREVIATIONS: NuRD, nucleosome remodeling deacetylase complex.
In some cancers, LSD1 can associate with various other transcriptional co-activators and co-repressors. For instance, in prostate cancers, LSD1 can interact with the androgen receptor to activate genes that drive tumor growth. This broad association indicates that inhibiting LSD1’s demethylation and scaffolding properties may have anti-tumor purposes across various cancer types.